Introduction
CRISPR genome editing is used to alter specific genomic DNA sequences, but the introduction of a specific mutation or gene requires an HDR donor template encoding the desired sequence. It can be challenging and time-consuming to create these templates for large insertions. In addition to these issues, some common donor formats, such as PCR amplicons, can integrate into a genome in unexpected ways. Here, we introduce a new donor template format with a faster manufacturing time and wider acceptance range for sequence complexities than previous IDT donor options, and that reduces many of the undesirable editing outcomes seen with other dsDNA donor options.
Alt-R HDR Donor Blocks are linear dsDNA fragments with a maximum length of 3 kb designed for use as donor templates in HDR-based, CRISPR knock-in experiments. Because they are manufactured using high-fidelity processes, they have a median error rate about 1 in 12,000 bases, which reduces the likelihood of incorporating an unintentional mutation in the genome being edited. To speed production and accept more complex sequences, IDT also adds universal sequences with proprietary chemical modifications on both ends of the donor template. The universal sequences have been rationally designed to facilitate efficient synthesis of the Alt-R HDR Donor Blocks, but do not themselves integrate into the target genome during HDR. The chemical modifications have been selected to help increase HDR frequency while blocking undesirable editing outcomes that can occur with unmodified, linear dsDNA templates. Below, we discuss the benefits of this approach as well as compare Alt-R HDR Donor Blocks to other commonly used donor template formats.
Comparison to other HDR donor options
| Donor format | Molecule type | Length | Business days to produce | Cost | HDR efficiency and fidelity |
|---|---|---|---|---|---|
| Genes in plasmid | dsDNA | 25–5000+ bp | 8–25+ | $$ | * |
| gBlocks™ Gene Fragments | dsDNA | 125–3000 bp | 2–8 | $ | * |
| Megamer™ Single-Stranded DNA Fragments | ssDNA | 200–2000 nt | 20+ | $$$ | ** |
| Alt-R HDR Donor Blocks | dsDNA | 200–2000 bp 2000–3000 bp |
12+ 18+ |
$$ | *** |
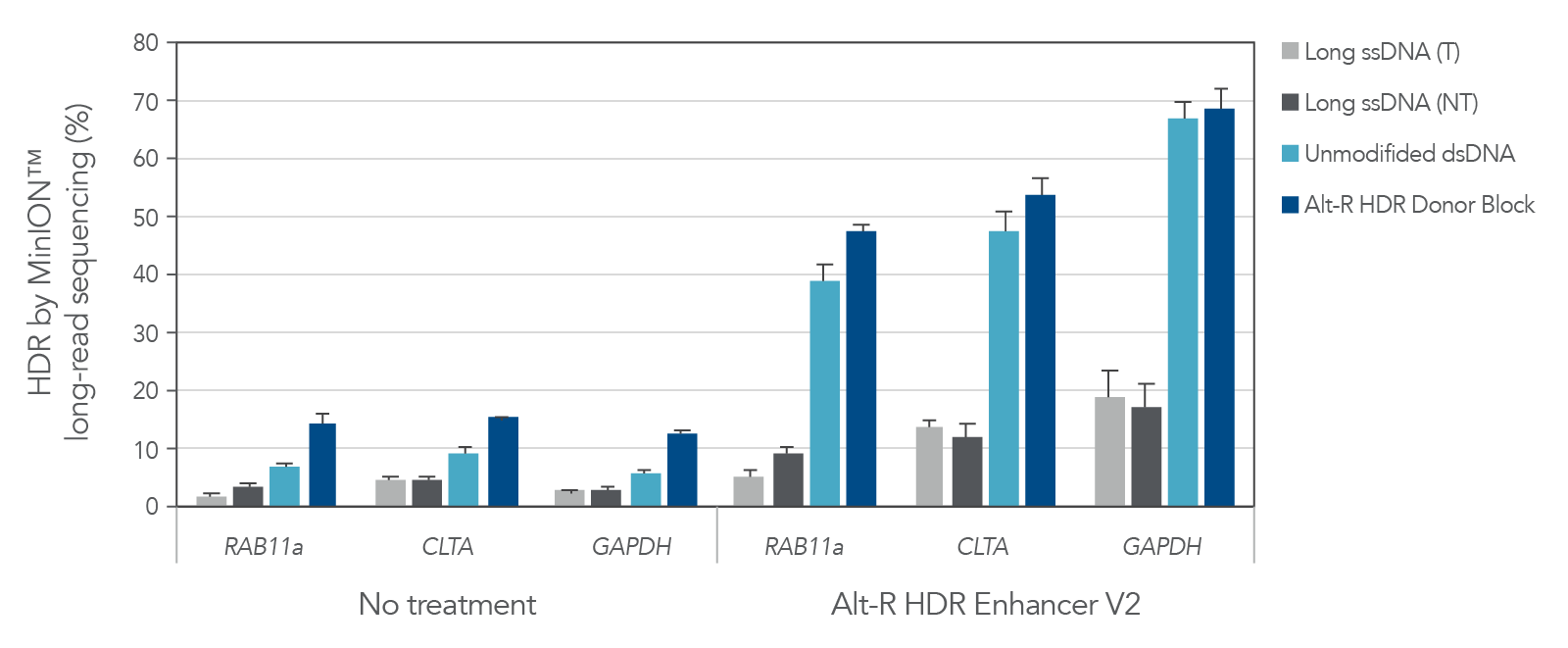
A variety of molecules are commonly used as donor templates once an experiment exceeds the 200 nt limit of our Ultramer™-based Alt-R HDR Donor Oligos. These include PCR amplicons, linearized plasmids, and large ssDNA constructs. At IDT we have offered linear dsDNA (gBlocks Gene Fragments) and long, linear ssDNA (Megamer Single-Stranded DNA Fragments) for some time, but until now have generally recommended ssDNA over dsDNA for HDR applications. A key metric of any HDR-based, gene knock-in experiment is the proportion of genomes that are correctly edited. Compared to other common donor formats, Alt-R HDR Donor Blocks have been shown to consistently result in the highest rates of HDR (Figure 1). Combining Alt-R HDR Donor Blocks with Alt-R HDR Enhancer V2 results in optimized HDR rates, with knock-in frequency as high as 70% as measured by MinION™ long-read sequencing in the following example.
Prior to the development of Alt-R HDR Donor Blocks, IDT recommended the use of long ssDNA donors (Megamer Single-Stranded DNA Fragments) to avoid the non-homologous integrations that are observed with unmodified, dsDNA donors. While it is still true that ssDNA does not incorporate into genomes in this way, recent publications as well as internal research data (Figure 2, lower panels, arrows) have shown that long ssDNA repair templates can produce partial, incomplete integration events at the on-target site. These truncated inserts can be difficult to assess in a pool of edited cells without long-read sequencing, but can occur at appreciable levels and appear as insertions with missing sequence corresponding to the 5’ end of the ssDNA donor. These types of mutations are not observed with dsDNA donor templates (Figure 2, top panel). The manufacturing process used for Alt-R HDR Donor Templates also allows for the acceptance of more complex sequences, a faster turnaround time, and higher yields when compared to Megamer Single-Stranded DNA Fragments.

Figure 2. Partial insertion of ssDNA observed via long read sequencing. Repair events at the CLTA locus following GFP knock-in were assessed by PacBio SMRT™ long-read sequencing. HDR aligned reads were visualized using Integrative Genomics Viewer (IGV) to assess integration rates across the full HDR insert. Individual repair events are represented as horizontal bars across each panel. While primarily complete HDR insertion events were observed for the dsDNA donors, use of long ssDNA donors resulted in partial HDR insertion events in which incomplete integration was observed at the 5’ junction of the ssDNA donor (arrows).
Reducing undesired non-homologous integration with dsDNA donors
Other factors beyond the overall HDR rate are also important to consider in long donor knock-in experiments. One of the main shortcomings of unmodified, linear dsDNA as a donor template is the ability of these molecules to integrate via non-homologous end joining (NHEJ). Since this process does not depend on homology between the donor and the targeted genomic site, NHEJ-mediated integration can occur at both the on-target cut site as well as any off-target cut sites of the CRISPR nuclease throughout the genome. While these types of insertions at the on-target site can easily be detected by sequencing, assessing off-target integration requires both the identification of the off-target loci as well as sequencing. The addition of chemically modified ends to Alt-R HDR Donor Blocks helps block non-homologous integration and reduce this undesired repair outcome compared to unmodified dsDNA. As shown in the following example, the chemical modifications reduced non-homologous integration at a mock off-target double-strand break (DSB) 3-fold relative to an unmodified dsDNA donor (Figure 3). Addition of Alt-R HDR Enhancer V2 pushes the balance of DNA repair pathways even further towards HDR and minimizes these blunt integration events. As a consequence of the chemical modifications, Alt-R HDR Donor Blocks are not recommended for knock-in approaches that rely upon NHEJ repair, such as homology-independent targeted insertion (HITI).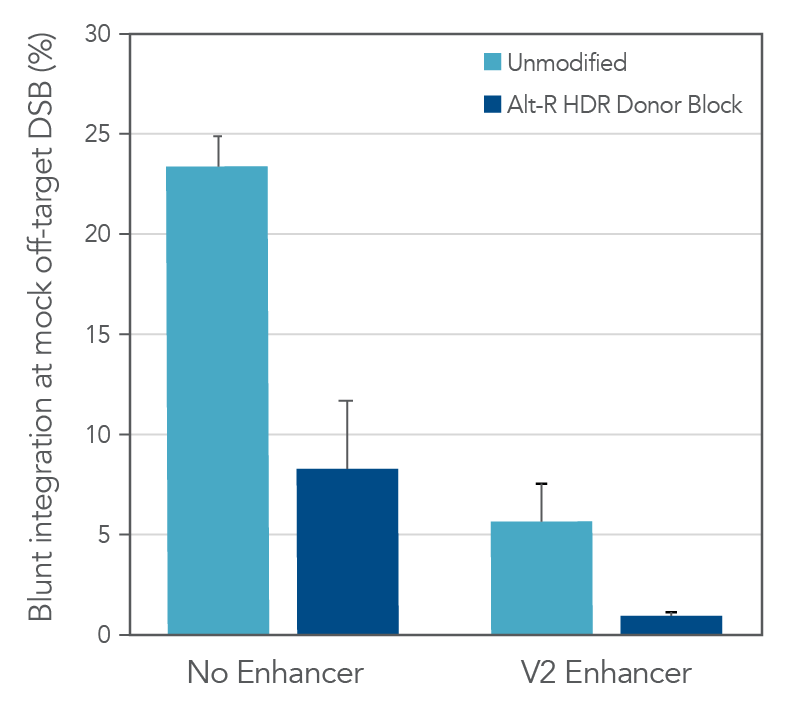
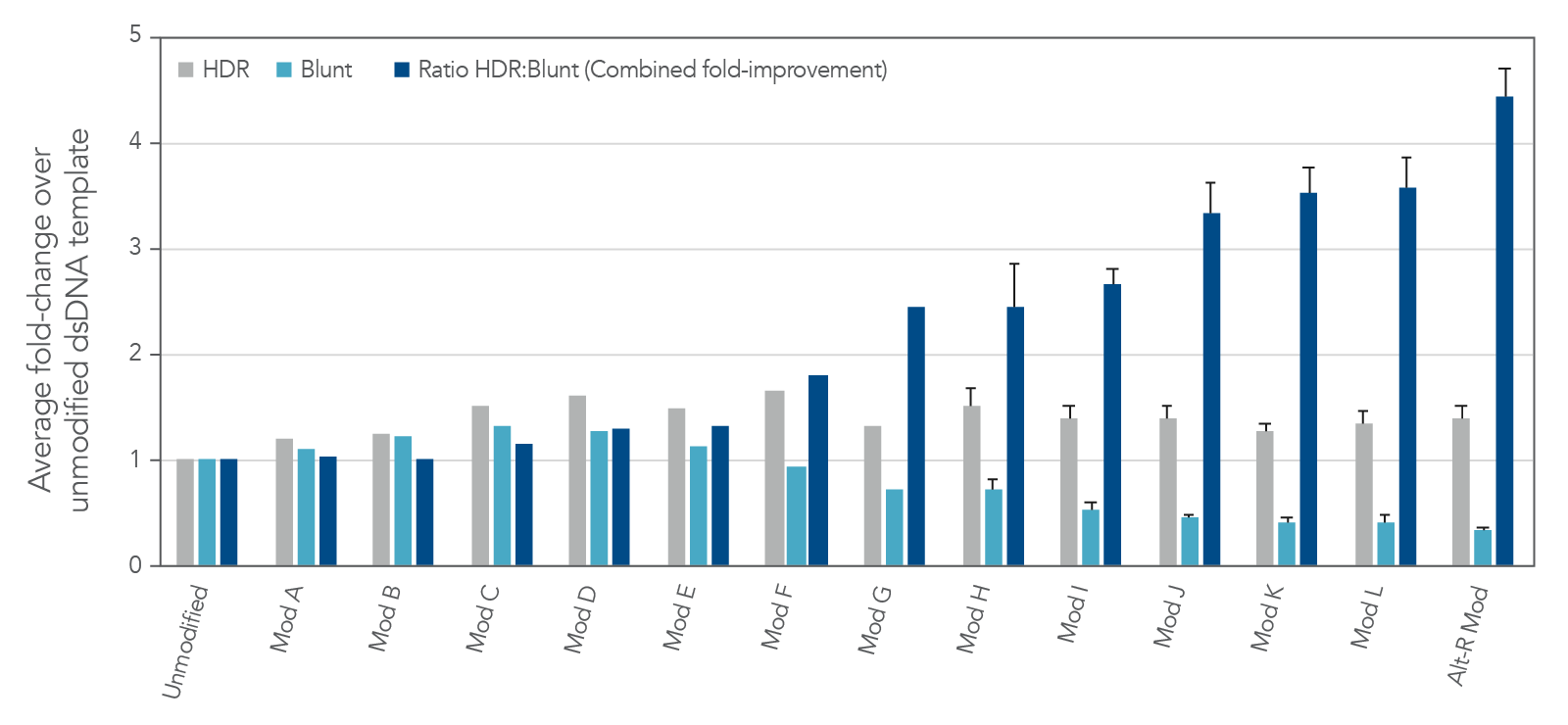
The modification pattern utilized in Alt-R HDR Donor Blocks are a result of careful screening of different modification patterns (Figure 4). IDT has identified a set of proprietary modifications that drive the editing outcomes towards HDR while mitigating the risk for blunt insertion when using dsDNA as the donor format.
To ensure these modifications are incorporated into the donor template consistently, regardless of the donor sequence itself, IDT appends universal sequences onto all Alt-R HDR Donor Blocks. These universal sequences allow IDT to accept more complex sequences for synthesis while decreasing synthesis time. Long read sequencing data have also demonstrated that these universal sequences are not incorporated by HDR and are not detectable in the genome following editing (as can be observed in Figure 2).
Design recommendations and protocols
The design of Alt-R HDR Donor Blocks follows similar rules to previous recommendations for Megamer Single-Stranded DNA Fragments. Homology arms should be 200–300 bp in length and symmetrical on the 5’ and 3’ ends if possible. In our experience, homology arms longer than 300 bp do not further increase HDR rates, nor do asymmetric homology arm lengths. The length of the arms may be decreased slightly if sequence complexity is present and interferes with synthesis of the donor. Maintaining at least 100 bp homology arms is recommended. Further recommendations on HDR Donor Block design can be found in our HDR application note. While it is possible to design effective pairs of gRNA and donor templates manually, IDT recommends scientists use our Alt-R HDR Design Tool. The tool automatically applies an extensive set of design rules based on empirical experimentation and has been updated to enable design of Alt-R HDR Donor Blocks instead of Megamer Single-Stranded DNA Fragments for large inserts.
Optimal conditions for delivering Alt-R HDR Donor Blocks alongside CRISPR ribonucleoprotein (RNP) complexes vary depending on the delivery method and cell type or system being used. IDT has developed protocols for common CRISPR delivery methods in cell culture and hosts several user-submitted protocols for other systems. Due to the recognition of cytosolic dsDNA by innate immune response pathways, the final concentration of Alt-R HDR Donor Block is often limited by the total mass of dsDNA. When adapting protocols for your application, be sure to titrate the concentration of the Alt-R HDR Donor Block carefully, to maximize HDR rates while limiting cytotoxicity.
Conclusion
The ability to integrate large, complex sequences into specific genomic coordinates using CRISPR is enabling entirely new areas of research while accelerating experiments that previously took weeks or months to complete. By enabling the fast, easy, and reliable production of large donor templates, Alt-R HDR Donor Blocks continue this trend of advancing CRISPR-based research. Combined with other tools from IDT such as Alt-R HDR Enhancer V2, scientists are now able to generate specific mutations faster and with higher confidence than ever before.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.