What is genotyping?
Overview
Genotyping is the process of determining the DNA sequence, called a genotype, at positions within the genome of an individual. Sequence variations can be used as markers in linkage and association studies to determine genes relevant to specific traits.

Identifying sequence variation
Each biological species is defined by a distinct set of common characteristics, but even within a species, there are subtle differences among individuals. In the human species, we can easily spot physical differences such as hair or eye color among people. In other species—even those considered non-living, such as viruses—certain differences among individuals also exist but they may be more subtle. Although these differences are not drastic enough to be called a distinct species, individuals within a species that do have slightly different characteristics are called variants.
Variation is the term used to describe the characteristic that is different within a population of distinct individuals or within a single species. For example, some populations of soybean plants are resistant to a common fungal pathogen, such as phytophthora root rot, whereas other populations are susceptible to this pathogen. In simplistic terms, fungal resistance is a variation. The phytophthora root rot-resistant population possesses a special feature enabling the plants to defend themselves against the fungus. If the fungal resistance is passed onto subsequent generations of plants, this feature is most likely coded in the plant’s genome.
Both environmental and genetic differences play a role in these visible or phenotypic features. Since environmental changes are not heritable, most researchers are interested in studying genetic variations that result in these physical differences. Genetic variation can be passed to the next generation and can increase or decrease the fitness of a given species.
Genotyping is the experimental procedure that identifies the differences in DNA sequence among individuals or populations. It is used to understand the connection between genotype (the underlying genetic code) and phenotype (the observable organismal structure). An individual genome is identified as a distinct variant when compared to a reference sequence, which is derived from the general population or a defined subgroup. A variant sequence can differ from the reference sequence in several ways. Types of genetic variation include single nucleotide variants (SNV), single nucleotide polymorphisms (SNPs), insertions and deletions (indels), translocations, and copy number variation (CNV).
SNPs are the most common type of sequence variant investigated by researchers; they are typically defined as SNVs that occur in >1% of the population. Based on the number of SNPs cataloged in the SNP database (dbSNP) [1] and a haploid genome size of 3.1 x 109 bp [2], the human genome should contain a SNP approximately once every 9.5 bases. Other common model organisms show a similarly high frequency of SNPs [3].
What is a haplotype?
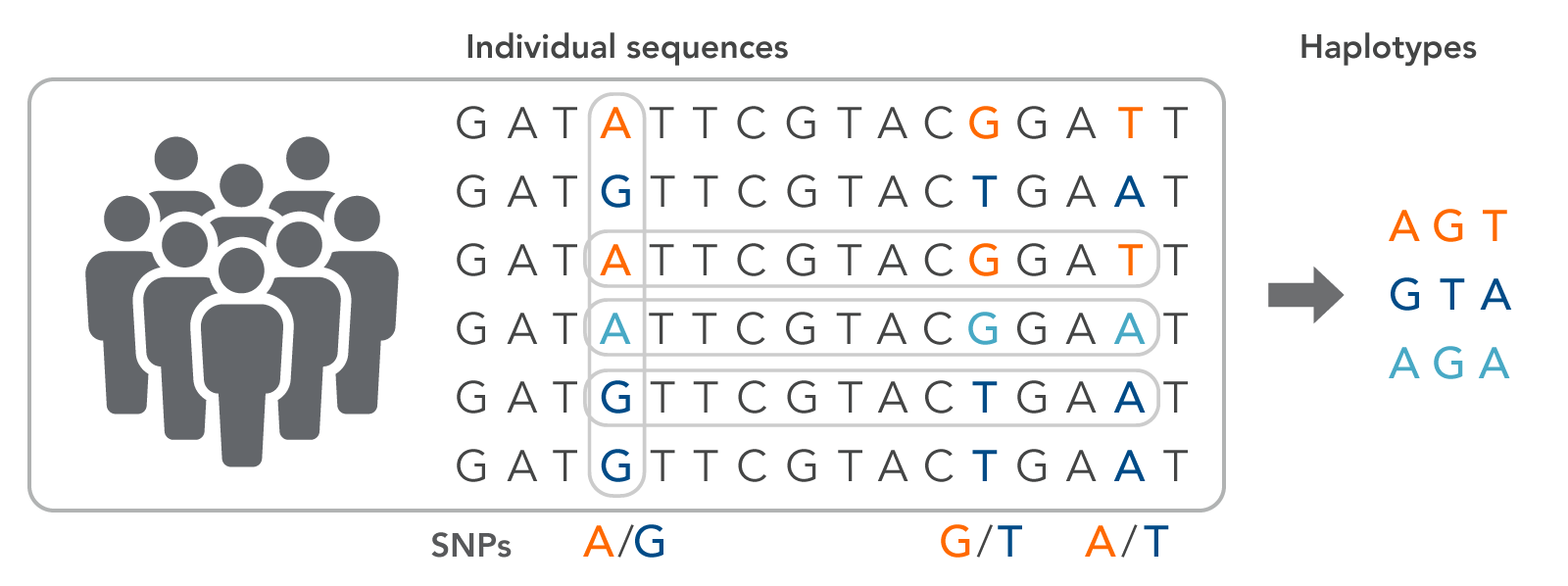
A haplotype is a series of adjacent SNPs from an individual that can serve as a signature for a specific phenotypic trait (Figure 1). The haplotype group, or block, is inherited from a single parent since individual genomic variants are located close together in the chromosome, which also means that haplotypes can be used to trace lineage. Haplotypes are often used to trace valuable traits for crop production, such as yield or disease resistance. Signature haplotypes may also indicate a predisposition to a specific disease or drug susceptibilities.
Associating sequence variation to specific traits
By comparing genetic variations among individuals of a species, researchers can identify heritable genetic signatures, or markers, relevant to specific traits. These unique differences can be used as markers in linkage and association studies.
Genome-wide association studies, or GWAS, compare genetic differences across entire genomes from two individuals or populations. For example, the genomes of a group of people that have a disease can be compared to genomic sequences from a similar group of people without the disease. Any SNP or haplotype that is more prevalent in those with the disease is called an associated genetic marker.
Association is merely the beginning, and many studies are needed to confirm if the genetic variation is truly the underlying genetic change that codes for the variant or trait. The National Center for Biotechnology Information (NCBI) maintains a registry of human genomic variations and their relationship to health called ClinVar. The website archives variations in human genes and the evidence supporting the association to a particular phenotype. Besides human health, finding the relationship of genotype to phenotype has many applications, such as:
- Personalized medicine
- Animal and plant breeding facilitation, such as the selection of a desired genotype from a cross hybrid
- Ancestry or disease tracing; map evolution, such as phylogenetic relationships between species; or inheritance or paternity testing
- Genotyping to identify a specific individual
- Pathogen typing and resistance screening
- Biodiversity monitoring
Technologies for SNP genotyping
There are varied methods to SNP genotyping depending on the number of samples, the number of genotypes to be tested, and the amount of sample material available, all factoring into the choice of technology.
High-throughput genotyping methods include whole genome analysis by NGS, SNP analysis using microarrays, and targeted sequencing methods such as amplicon sequencing or hybridization capture technology.
Low-throughput analyses include using multiplex quantitative PCR (qPCR), PACE™ (PCR Allele Competitive Extension) SNP genotyping, and multiplex digital PCR (dPCR) to identify the genotype of a specific SNP.
IDT genotyping methods
Genotyping by qPCR
Quantitative polymerase chain reaction (qPCR) is a commonly used genotyping technique. Often employing a primer-pair and target-specific fluorescent probe, qPCR can be a high-resolution way to identify SNPs. IDT offers a complete SNP genotyping solution with predesigned sequences, as well as complementary easy- and ready-to-use reagent mixes. For other applications, modified probes are available that can be incorporated into custom qPCR assays.
Targeted next generation sequencing
Targeted next generation sequencing (NGS) focuses on specific regions of interest in the genome. With targeted NGS, researchers can target specific genes, coding regions, or even chromosomal segments at deeper coverage than alternative sequencing methods, obtaining fast, accurate, and precise genomic insights.
PACE™ SNP Genotyping Assays
PACE (PCR Allele Competitive Extension) SNP genotyping assays are well-suited for projects that require a reliable, cost-effective approach for SNP and indel identification.
Genotyping by digital PCR (dPCR)
Digital PCR (dPCR) includes the same reagents found in a typical qPCR assay and amplified in a similar manner, but dPCR divides the reaction into nano-sized droplets or wells before amplification. The partitions are so small that either 1 or 0 templates are in each. After amplification, the fluorescence in the well or droplet represents the genotype.
Get started with genotyping
For user-defined methods, order custom probes with modifications, such as Affinity Plus™ qPCR locked nucleic acid probes, that increase probe hybridization affinity and enable designs within difficult sequences and with selective target identification.
xGen NGS Amplicon Sequencing
Our amplicon solutions enable you to advance from sample to sequencing faster, without sacrificing coverage or yields. If your research requires rapid, ready-to-sequence materials or you are working with low-input quantities, xGen NGS can unlock the answers you seek.
Affinity Plus™ qPCR Probes
Use Affinity Plus qPCR Probes for enhanced discrimination of thermodynamically similar samples such as single nucleotide polymorphisms and transcript variants. The Affinity Plus bases used in these qPCR probes include up to 6 locked nucleic acid monomers. When incorporated into a probe, locked nucleic acids impart heightened structural stability, leading to increased hybridization melt temperature (Tm).
PACE™ SNP Genotyping Assays
PACE (PCR Allele Competitive Extension) SNP genotyping assays are well-suited for projects that require a reliable, cost-effective approach for SNP and indel identification.
PACE is a trademark of 3CR Bioscience
rhAmp™ SNP Genotyping System
The rhAmp SNP Genotyping System is a fully integrated genotyping solution that includes an extensive predesigned sequence collection, a custom design tool, reagent mixes, and optional synthetic control templates. SNP identification may be performed on any commonly available qPCR instrument.
References
- dbSNP Build 151 NCBI 2017. www.ncbi.nlm.nih.gov/dbvar/content/org_summary/. Accessed Apr 14, 2021.
- Genome Reference Consortium Human Build 38 patch release 13 (CRCh38.p13) NCBI 2019. ncbi.nlm.nih.gov/assembly/GCF_000001405.39/ Accessed Apr 14, 2021.
- Prediger E. Consider SNPs when designing PCR and qPCR assays. 2017. Accessed Jan 6, 2020.